| Name: | Chengyuan WANG | Gender: | |
|---|---|---|---|
| Education: | 2010-2016, Ph.D., Institute of Plant Physiology and Ecology, CAS, Shanghai, China 2006-2010, B.S., School of Life Science and Biotechnology, Shanghai Jiao Tong University, Shanghai, China |
||
| Academic degree: | Ph.D. | Academic title: | Principal Investigator |
| Departments: | Bacterial transcription regulation and antibiotics discovery | Discipline: | Structural Biology |
| Phone: | E-mail: | cywang@ips.ac.cn | |
| Mailing Address: | Life Science Research Building, 320 Yueyang Road, Xuhui District, 200031 | ||
| Curriculum vitae: |
|---|
2021-present, Principal investigator, Institut Pasteur of Shanghai, CAS, Shanghai, China |
| Research direction: |
|---|
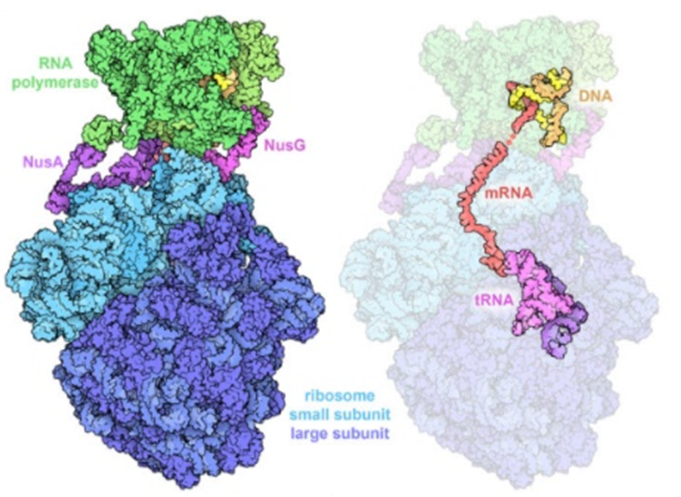
Transcription is the first and the most-regulated step of gene expression, by which RNA polymerase (RNAP) copies genetic information from DNA to RNA. It is fundamentally important to understand the structure, function, and regulation of the transcription machinery--RNAP. RNAPs are well conserved across bacterial species but are less conserved between bacteria and human. Therefore, bacterial RNAPs are promising drug target for antibiotic discovery. Our lab focusses on the molecular mechanism of transcription and transcription regulation, especially the mechanism of transcription and translation coupling. We also develop new antibiotics targeting different pathogenic bacterial RNAP. We employ multiple approaches in Structure biology (X-ray crystallography and cryo-EM), Biochemistry, Enzymology, Microbiology, and Bioinformatics.
|
| Research progress: |
|---|
|
Selected Publications (#co-first author) 1. Wang, C.Y.#, Molodtsov V.#, Firlar E., Kaelber J.T., Blaha G., Su M., & Ebright R. H. (2020). "Structural basis of transcription-translation coupling." Science 369(6509): 1359-1365. 2. Wang, C.Y.#, Li, J.#, Ma M., Zhu L., Lin W., & Zhang P. (2020). " Structural and biochemical insights into two BAHD acyltransferases (AtSHT and AtSDT) involved in phenolamide biosynthesis", Front. Plant. Sci. doi: 10.3389/fpls.2020.610118 3. Wang, H., Wang, C.Y., Fan, W., Yang, J., Appelhagen, I., Wu, Y., & Zhang, P. (2018). A novel glycosyltransferase catalyses the transfer of glucose to glucosylated anthocyanins in purple sweet potato. J Exp Bot. doi:10.1093/jxb/ery305 4. Li, J.#, Wang, C.Y.#, Yang, G., Sun, Z., Guo, H., Shao, K., Zhang, P. (2017). Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria. Proc Natl Acad Sci, 114(31), 8235-8240. doi:10.1073/pnas.1620183114 5. Zhou, F.#, Wang, C.Y.#, Gutensohn, M.#, Jiang, L., Zhang, P., Zhang, D., & Lu, S. (2017). A recruiting protein of geranylgeranyl diphosphate synthase controls metabolic flux toward chlorophyll biosynthesis in rice. Proc Natl Acad Sci, 114(26), 6866-6871. doi:10.1073/pnas.1705689114 6. Wang, C.Y.#, Chen, Q.#, Fan, D., Li, J., Wang, G., & Zhang, P. (2016). Structural Analyses of Short-Chain Prenyltransferases Identify an Evolutionarily Conserved GFPPS Clade in Brassicaceae Plants. Mol Plant, 9(2), 195-204. doi: 10.1016/j.molp.2015.10.010 7. Zhao, Q., Wang, C., Wang, C.Y., Guo, H., Bao, Z., Zhang, M., & Zhang, P. (2015). Structures of FolT in substrate-bound and substrate-released conformations reveal a gating mechanism for ECF transporters. Nature Commun, 6, 7661. doi:10.1038/ncomms8661 8. Qi, X., Lin, W., Ma, M., Wang, C.Y., He, Y., He, N., Zhang, P. (2016). Structural basis of rifampin inactivation by rifampin phosphotransferase. Proc Natl Acad Sci, 113(14), 3803-3808. doi:10.1073/pnas.1523614113 9. Yu, F., He, F., Yao, H., Wang, C.Y., Wang, J., Li, J., Zhang, P. (2015). Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex. EMBO Rep, 16(7), 813-823. doi:10.15252/embr.201540137 10. Lin, W., Wang, Y., Han, X., Zhang, Z., Wang, C.Y., Wang, J., Zhang, P. (2014). Atypical OmpR/PhoB subfamily response regulator GlnR of actinomycetes functions as a homodimer, stabilized by the unphosphorylated conserved Asp-focused charge interactions. J Biol Chem, 289(22), 15413-15425. doi:10.1074/jbc.M113.543504 11. Xu, K., Zhang, M., Zhao, Q., Yu, F., Guo, H., Wang, C.Y., Zhang, P. (2013). Crystal structure of a folate energy-coupling factor transporter from Lactobacillus brevis. Nature, 497(7448), 268-271. doi:10.1038/nature12046 |
| Laboratory members: |
|---|